Nils Eckstein
Machine Learning Research Scientist
CV
Contact
About Me
Hi, I am Nils. I am a machine learning researcher working on DNN interpretability. Before that I was doing my PhD at ETH Zurich and HHMI Janelia Research Campus where I worked on the development of computer vision algorithms for the analysis of large scale biological image datasets, in particular the recently imaged fruit fly brain (zoom in to see
individual neurons, mitochondria and other structures). Before becoming fascinated by (real & artificial) brains, I studied physics and worked on simulating the supermassive active black holes in the center of galaxies. Besides that I like to make music, dabble in generative art, and think about how to become immortal. I believe that we might be the last generation of humans that still have to die. Depressing, right?
Projects
Microtubule Tracking in large scale EM datasets
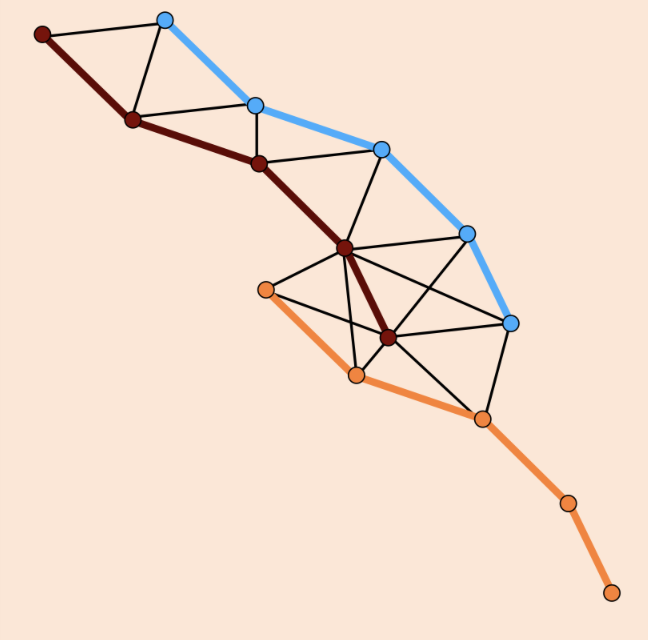
 A method and library for automatic tracking of microtubules in terabyte
scale EM datasets. This is made possible by combining deep learning
and discrete optimisation, exploiting priors on microtubule structure. We managed to increase tracking accuracy by more than 50% and propose a highly efficient ILP formulation that leads to 4 orders of magnitude faster solve times compared to a naive encoding. Code.
A method and library for automatic tracking of microtubules in terabyte
scale EM datasets. This is made possible by combining deep learning
and discrete optimisation, exploiting priors on microtubule structure. We managed to increase tracking accuracy by more than 50% and propose a highly efficient ILP formulation that leads to 4 orders of magnitude faster solve times compared to a naive encoding. Code.
Neurotransmitter Classification
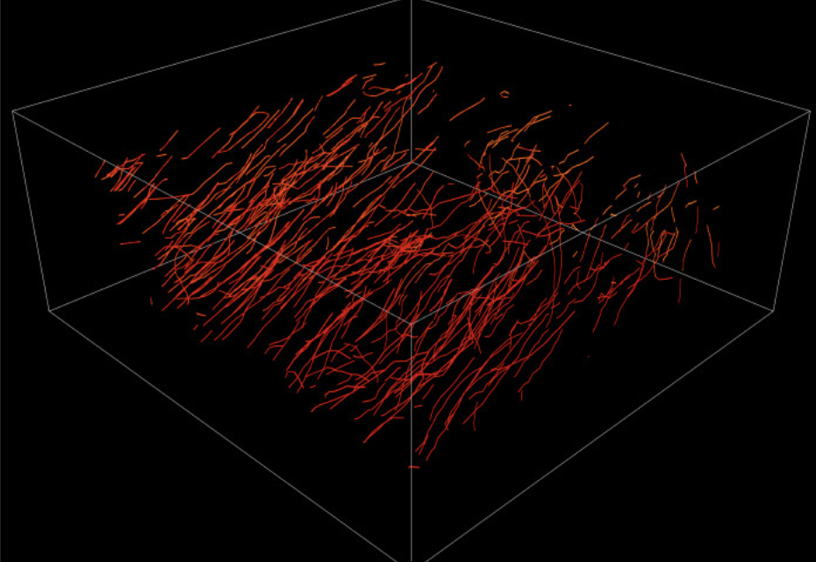
 A surprising finding that deep neural networks are able to predict the neurotransmitter of a given presynaptic site by looking at electron microscopy images of the synaptic site alone. A task that humans cannot generally perform in the fruit fly brain. Code.
A surprising finding that deep neural networks are able to predict the neurotransmitter of a given presynaptic site by looking at electron microscopy images of the synaptic site alone. A task that humans cannot generally perform in the fruit fly brain. Code.
Discriminative Attribution from Counterfactuals
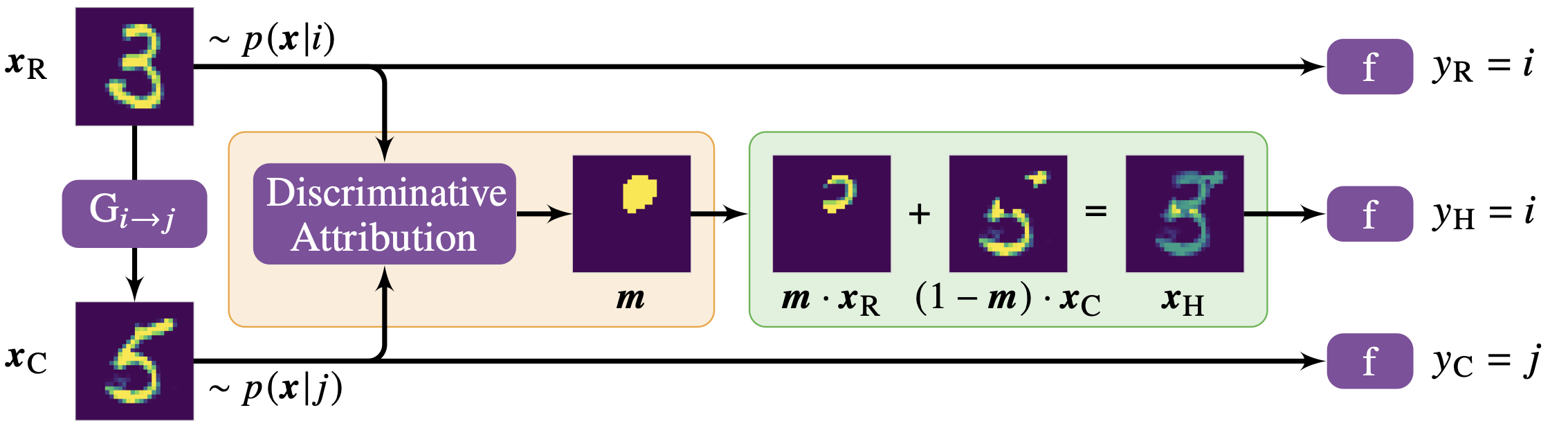 A novel method for DNN interpretability that allows humans to understand subtle class differences as learned by an image classifier. We used it to uncover so far unknown phenotypical differences in synapses that express different neurotransmitters. Code.
A novel method for DNN interpretability that allows humans to understand subtle class differences as learned by an image classifier. We used it to uncover so far unknown phenotypical differences in synapses that express different neurotransmitters. Code.
Whole Cell Organelle Segmentation
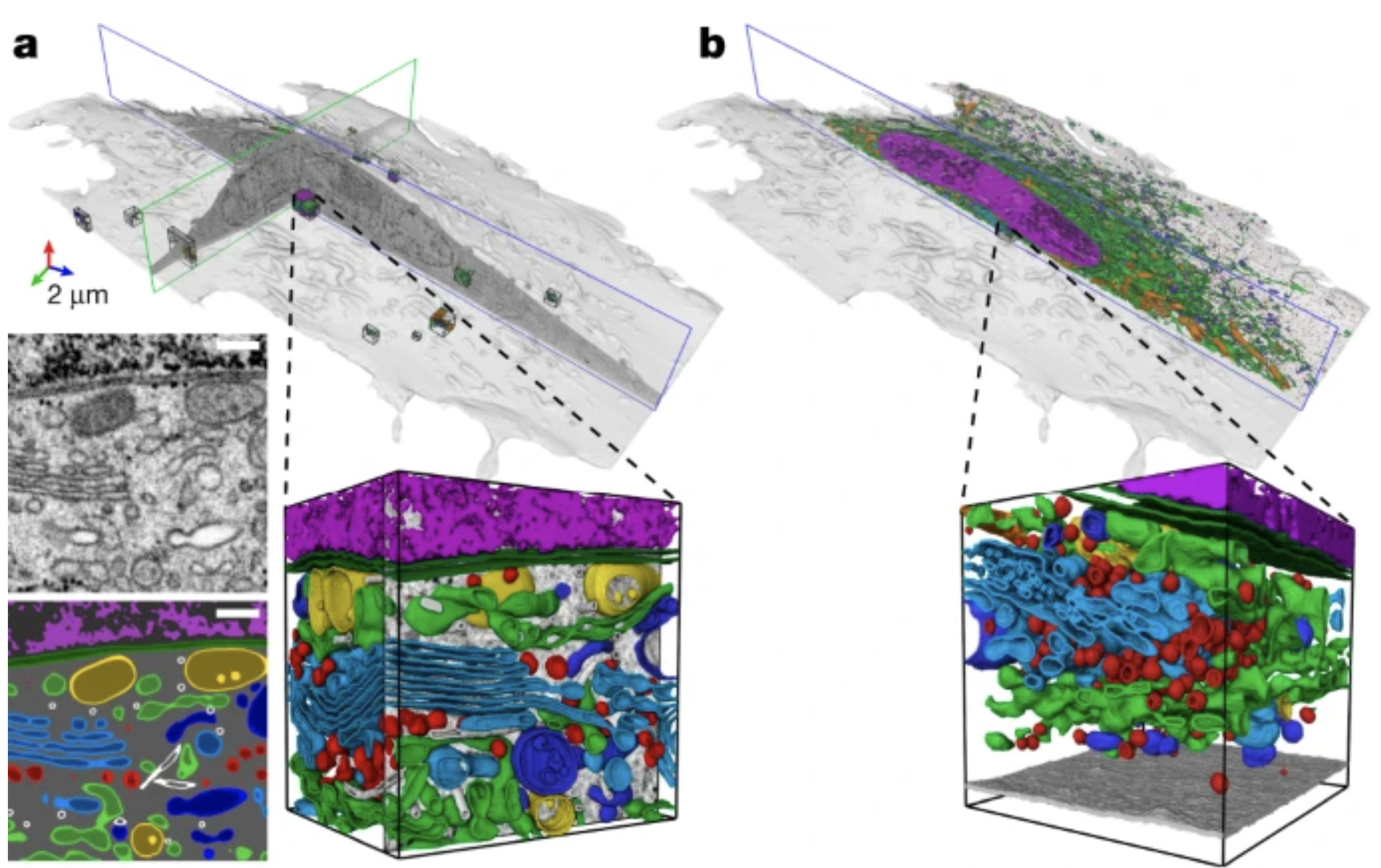 I helped segmenting nearly all organelles in super high resolution isotropic EM datasets, resulting in probably the most accurate map of a cell we have to date. You can browse through the data here.
I helped segmenting nearly all organelles in super high resolution isotropic EM datasets, resulting in probably the most accurate map of a cell we have to date. You can browse through the data here.
Reverse game of life
 For this website I was interested in picking a start state for the game of life that eventually converges to a message or an image. Unfortunately the game of life is irreversible as information is destroyed constantly. However, we can still search for pre-images of a given state that will lead to the target state after one step of evolution. This library utilizes integer linear programming to find a valid pre-image of a game of life state.
For this website I was interested in picking a start state for the game of life that eventually converges to a message or an image. Unfortunately the game of life is irreversible as information is destroyed constantly. However, we can still search for pre-images of a given state that will lead to the target state after one step of evolution. This library utilizes integer linear programming to find a valid pre-image of a game of life state.
A strange model of computation - Integer Gradient Descent
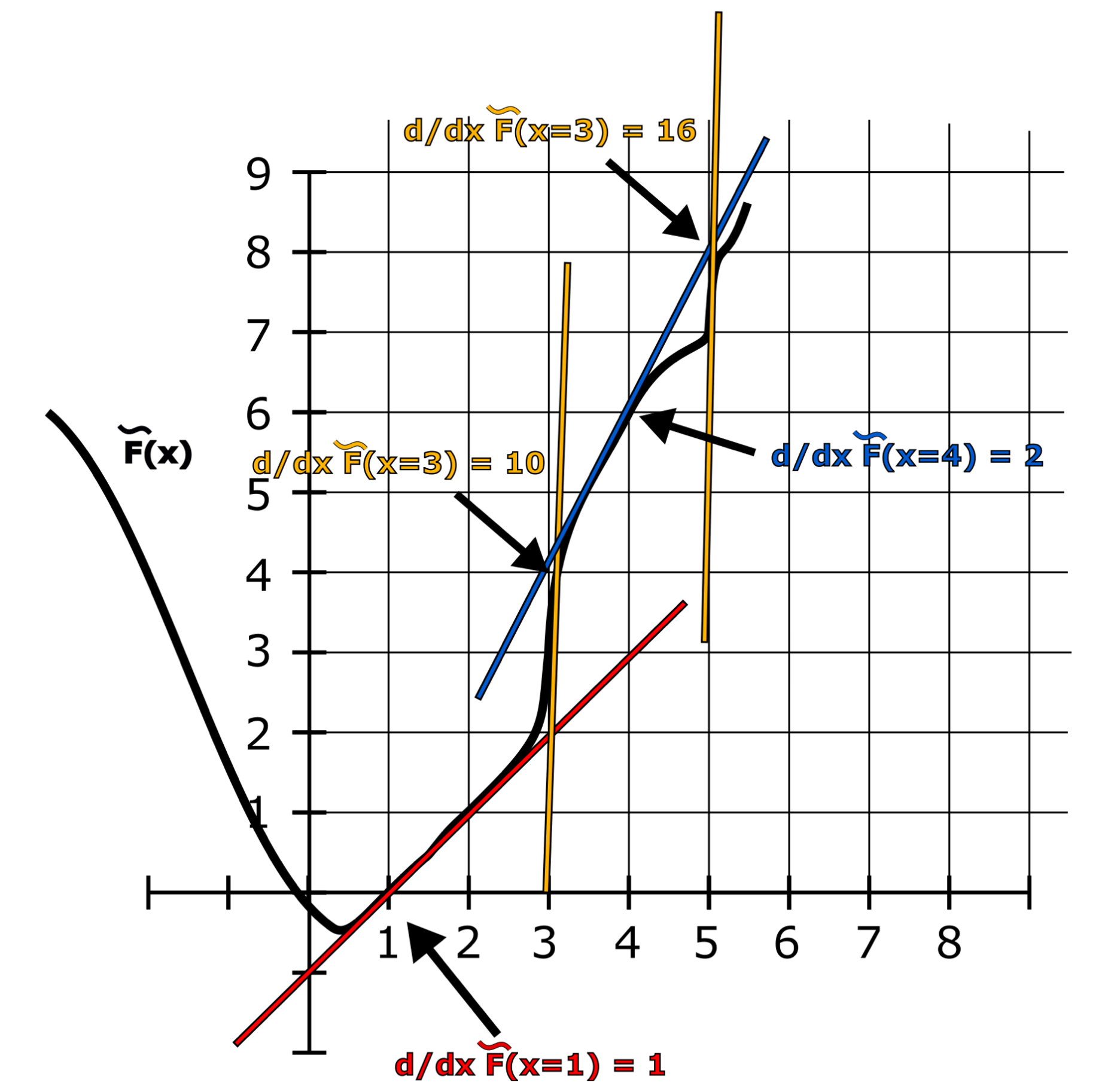 An esoteric model of computation that can be understood as a generalization of FRACTRANs and used to formulate an analytical version of the Collatz conjecture. It is also Turing complete.
An esoteric model of computation that can be understood as a generalization of FRACTRANs and used to formulate an analytical version of the Collatz conjecture. It is also Turing complete.
Art
Curious Things
About Me
Hi, I am Nils. I am a machine learning researcher working on DNN interpretability. Before that I was doing my PhD at ETH Zurich and HHMI Janelia Research Campus where I worked on the development of computer vision algorithms for the analysis of large scale biological image datasets, in particular the recently imaged fruit fly brain (zoom in to see individual neurons, mitochondria and other structures). Before becoming fascinated by (real & artificial) brains, I studied physics and worked on simulating the supermassive active black holes in the center of galaxies. Besides that I like to make music, dabble in generative art, and think about how to become immortal. I believe that we might be the last generation of humans that still have to die. Depressing, right?
Projects
Microtubule Tracking in large scale EM datasets

Neurotransmitter Classification

Discriminative Attribution from Counterfactuals

Whole Cell Organelle Segmentation

Reverse game of life
A strange model of computation - Integer Gradient Descent